Figures & data
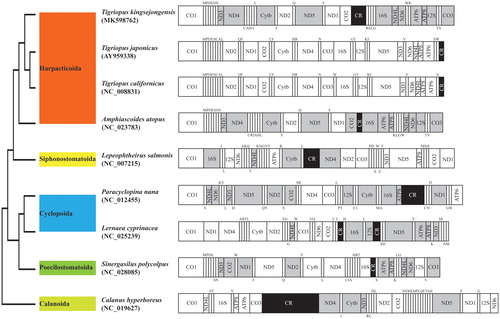
Figure 1. Phylogenetic analysis. We conducted comparison of the mitochondrial genomes of nine copepods. Amino acids of 13 PCGs gene from nine copepods were aligned using MEGA software (ver. 10.0.1) with the ClustalW alignment algorithm. To establish the best-fit substitution model for phylogenetic analysis, the model with the lowest Bayesian Information Criterion (BIC) and Akaike Information Criterion (AIC) scores were estimated using a maximum likelihood (ML) analysis. According to the results of model test, maximum likelihood phylogenetic analyses were performed with the GTR + G+I model. Gray, the opposite direction of PCGs; Black, control regions; Upper and lower single letters indicate tRNA clockwise and anticlockwise, respectively.