Figures & data
Figure 1. Eastern cottontail (EC), New England cottontail (NEC), and snowshoe hare (SSH) samples identified in 5 Northeastern states from 2003 to 2012, by county. Samples with unknown county locations are as follows: 9/14/0 for Connecticut, 1/15/0 for Massachusetts, 0/0/2 for New Hampshire, and 36/0/0 for Rhode Island for EC/NEC/SSH, respectively.
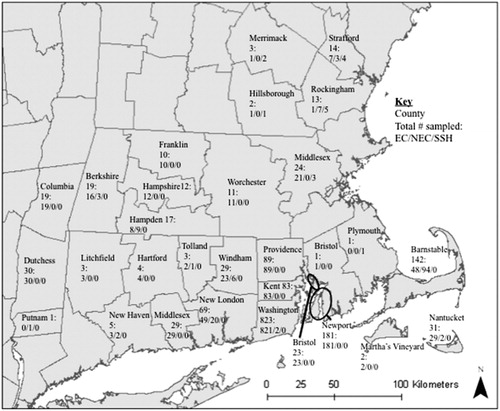
Figure 2. Mitochondrial DNA control region nucleotides used for specimen identification to distinguish among three lagomorph species: eastern cottontail (Sylvilagus floridanus), New England cottontail (S. transitionalis), and snowshoe hare (Lepus americanus). Highlighted nucleotides are diagnostic characters (DC) and vertical numbers above nucleotide position indicate their position in the sequence alignment inclusive of all haplotypes, relative to the reference sequence (GenBank accession: NC024043) starting at position 15357 and ending at 15895. A – indicates a gap in the alignment. Only diagnostic characters are shown here. Supplemental Figure 1 should be used for sequence alignment for species identification.

Supplemental Material
Download Zip (481.2 KB)Data accessibility
GenBank accession numbers are KC23298-KC923420 and all samples, except skull samples used for barcode testing, are archived at the University of Rhode Island’s Wildlife Genetics and Ecology Laboratory.
