Figures & data
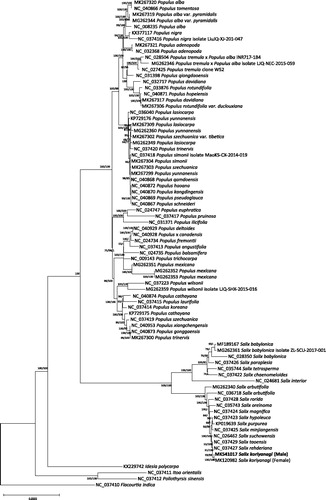
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 80 complete chloroplast genomes from Salicaceae: Salix koriyanagi (MK541017 in this study and MK120982), Salix suchowensis (NC_026462), Salix purpurea (KP019639), Salix rehderiana (NC_037427), Salix rorida (NC_037428), Salix taoensis (NC_037429), Salix tetrasperma (NC_035744), Salix paraplesia (NC_037426), Salix oreinoma (NC_035743), Salix minjiangensis (NC_037425), Salix magnifica (NC_037424), Salix interior (NC_024681), Salix hypoleuca (NC_037423), Salix chaenomeloides (NC_037422), Salix babylonica (NC_028350, MG262361, and MF189167), Salix arbutifolia (NC 036718 and MG262340), Populus yunnanensis (MG262360 and MK267299), Populus xiangchengensis (NC_040953), Populus × canadensis (NC_040928), Populus wilsonii (MG262359 and NC_037223), Populus trinervis (NC_037420 and MK267300), Populus trichocarpa (NC 009143), Populus tremula × Populus alba (MG262346 and NC_028504), Populus tremula (NC_027425), Populus tomentosa (NC_040866), Populus szechuanica var. tibetica (MK267302), Populus szechuanica (NC_037419 and MK267303), Populus simonii (NC_037418 and MK267304), Populus schneideri (NC_040867), Populus rotundifolia var. duclouxiana (MK267306), Populus rotundifolia (NC_033876), Populus qiongdaoensis (NC_031398), Populus qamdoensis (NC_040868), Populus pseudoglauca (NC_040869), Populus pruinosa (NC_037417), Populus nigra (NC_037416 and KX377117), Populus mexicana (MG262351, MG262352, and MG262353), Populus laurifolia (NC_037415), Populus lasiocarpa (NC_036040, MG262349, and MK267309), Populus koreana (NC_037414), Populus kangdingensis (NC_030870), Populus ilicifolia (NC_031371), Populus hopeiensis (NC_040871), Populus haoana (NC_040872), Populus gonggaensis (NC_040873), Populus fremontii (NC_024734), Populus euphratica (NC_024747), Populus deltoides (NC_040929), Populus davidiana (NC_032717 and MK267317), Populus cathayana (NC_040874), Populus balsamifera (NC_024735), Populus angustifolia (NC_037413), Populus alba var. pyramidalis (MG262344 and MK267319), Populus alba (NC_008235 and MK267320), Populus adenopoda (NC_032368 and MK267321), Populus cathayana (KP729175), Populus yunnanensis (KP729176), Poliothyrsis sinensis (NC_037412), Itoa orientalis (NC_037411), Idesia polycarpa (KX229742), and Flacourtia indica (NC_037410). Neighbor joining tree was used for displaying phylogenetic tree. The numbers above branches indicate bootstrap support values of neighbor joining maximum likelihood trees, respectively.