Figures & data
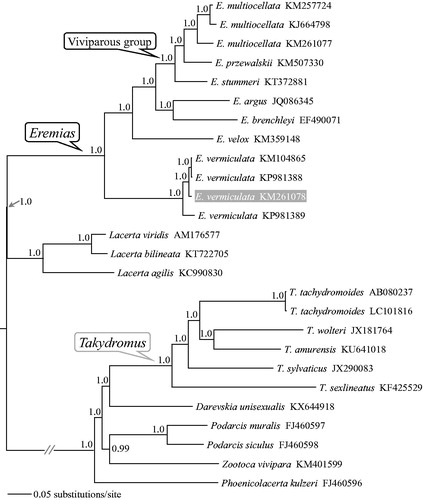
Figure 1. A majority-rule consensus tree derived from Bayesian inference using MrBayes v.3.2.2 (Ronquist et al. Citation2012) with GTR + GAMMA substitution model, based on the concatenated PCGs of 12 individuals of Eremias lizards and 14 outgroups. The tree was rooted using midpoint rooting method and the newly sequenced sample was highlighted in gray. GenBank accession numbers are given with species names. DNA sequences were aligned in MEGA v.7.0.14 (Kumar et al. Citation2016). The PCGs were translated into amino acid sequences and were manually concatenated into a single nucleotide dataset (in total 11,403 bp). Node numbers show Bayesian posterior probabilities. Branch lengths represent means of the posterior distribution.