Figures & data
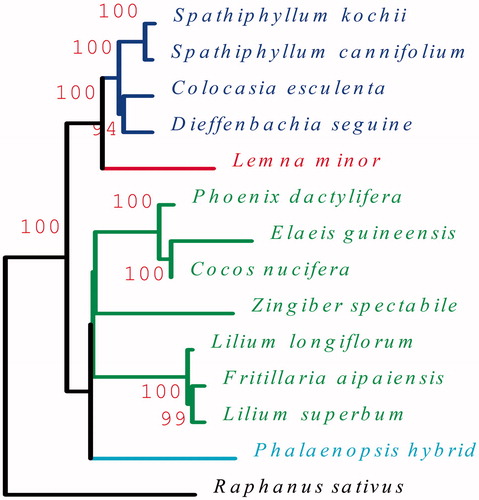
Figure 1. The ML phylogenetic tree of S. cannifolium and other species based on 24 protein-coding genes from 14 plant taxa. Raphanus sativus (NC_024469) was set as outgroup. Genbank accession numbers for each plant are: Colocasia esculenta (NC_016753), Dieffenbachia seguine (NC_027272), Spathiphyllum cannifolium (MK372232), Spathiphyllum kochii (KR270822), Lemna minor (NC_010109), Phalaenopsis hybrid (NC_007499), Cocos nucifera (KF285453), Elaeis guineensis (JF274081), Phoenix dactylifera (GU811709), Lilium longiflorum (KC968977), Lilium superbum (NC_026787), Fritillaria taipaiensis (NC_023247), Zingiber spectabile (NC_020363).