Figures & data
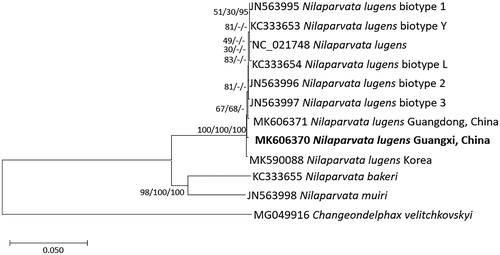
Figure 1. Neighbor joining (bootstrap repeat: 10,000), minimum evolution (bootstrap repeat: 10,000), and maximum likelihood (bootstrap repeat: 1,000) phylogenetic trees of 11 Delphacidae and one Fulgoridae complete mitochondrial genomes: Eight Nilaparvata lugens (MK606370 in this study, MK606371, MK590088, JX880069, JN563995, KC333653, JN563996, JN563997, and KC333654), Nilaparvata muiri (JN563998), Nilaparvata bakeri (KC333655), and Changeondelphax velitchkovskyi (MG049916) as an outgroup species. Phylogenetic tree was shown based on neighbor joining tree. The numbers above branches indicate bootstrap support values of neighbor joining, minimum evolution, and maximum likelihood phylogenetic trees, respectively.