Figures & data
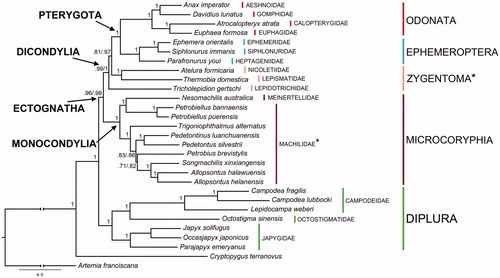
Figure 1. Bayesian phylogeny based on the concatenated set of 13 mitochondrial protein-encoding genes of the following basal hexapod lineages used in this study: Campodea fragilis DQ529236, Campodea lubbocki DQ529237 and Lepidocampa weberi JN990601 (Campodeidae; Diplura); Octostigma sinensis JN990598 (Octostigmatidae; Diplura); Japyx solifugus NC007214, Occasjapyx japonicus JN990600 and Parajapyx emeryanus JN990599 (Japygidae; Diplura); Allopsontus helanensis KJ754501, Allopsontus halawuensis KJ754500, Nesomachilis australica NC006895, Pedetontinus luanchuanensis KJ754502, Pedetontus silvestrii EU621793, Petrobiellus bannaensis KJ754503, Petrobiellus puerensis KJ754504, Petrobius brevistylis NC007688, Songmachilis xinxiangensis NC021384 and Trigoniophthalmus alternatus EU016193 (Microcoryphia); Atelura formicaria EU084035, Thermobia domestica AY639935 and Tricholepidion gertschi AY191994 (Zygentoma); Ephemera orientalis NC012645, Parafronurus youi NC011359 and Siphlonurus immanis NC013822 (Ephemeroptera); Atracalopteryx atrata NC027181 and Eupohaea formosa NC014493 (Zygoptera; Odonata); Anax imperator NC031821 and Davidius lunatus NC012644 (Anisoptera; Odonata). The collembolan Cryptopygus terranovus NC037610 and the crustacean Artemia franciscana NC001620 have been used as outgroups. Double branch-support on nodes (i.e. when different among alternative data sets) refers to analyses based on complete set of nucleotide codon positions/first and second only, respectively. *refers to paraphyletic groups.