Figures & data
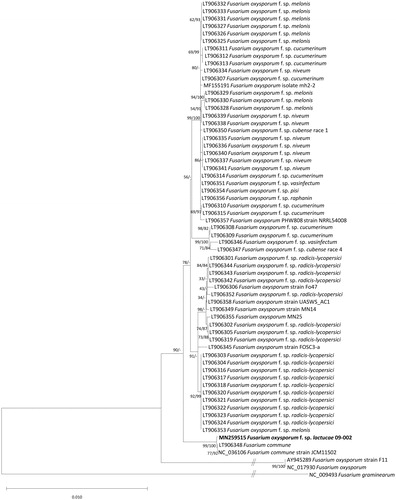
Figure 1. Neighbor joining and maximumlikelihood phylogenetic trees (bootstrap repeat is 10,000 and 1,000, respectively) of 64 Fusarium mitochondrial genomes: F. oxysporum f. sp. lactucae (MN259515 in this study), F. oxysporum (NC_017930, AY874423, AY945289, MF155191, LT906306, LT906349, LT906355, LT906358, LT906357, and LT906345), F. oxysporum f. sp. melonis (LT906328, LT906329, LT906330, LT906325, LT906326, LT906327, LT906331, LT906332, LT906333, and LT906353), F. oxysporum f. sp. pisi (LT906354), F. oxysporum f. sp. vasinfectum (LT906351), F. oxysporum f. sp. raphanin (LT906356), F. oxysporum f. sp. cubense race 1 (LT906350), F. oxysporum f. sp. cubense race 4 (LT906347), F. oxysporum f. sp. niveum (LT906334, LT906338, LT906335, LT906336, LT906340, LT906341, LT906339, and LT906337), F. oxysporum f. sp. cucumerinum (LT906315, LT906314, LT906310, LT906307, LT906308, LT906309, LT906311, LT906312, and LT906313), F. oxysporum f. sp. radicis-lycopersici (LT906352, LT906342, LT906343, LT906344, LT906302, LT906305, LT906319, LT906301, LT906303, LT906304, LT906316, LT906317, LT906318, LT906320, LT906321, LT906322, LT906323, and LT906324), F. oxysporum f. sp. vasinfectum (LT906346), F. commune (LT906348 and NC_036106), and F. graminearum (NC_009493) as an outgroup. Phylogenetic tree was drawn based on neighborjoining tree. Numbers on branches indicate bootstrap values of neighbor joining and maximum likelihood phylogenetic trees, respectively.