Figures & data
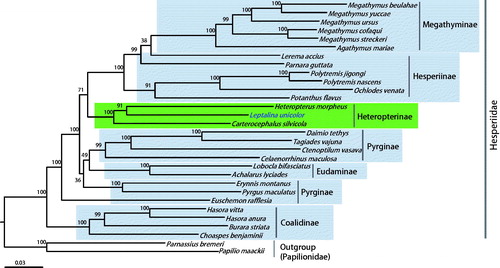
Figure 1. Phylogenetic tree of Hesperiidae, using maximum-likelihood (ML) method based on concatenated 13 protein-coding genes (PCGs) and 2 rRNAs. The numbers at each node specify bootstrap percentages of 1000 pseudoreplicates. Papilionidae (Papilio maakii and Parnassius bremeri) were utilized as the outgroup. GenBank accession numbers are as follows: Pyrgus maculatus, KP689265 (unpublished); Euschemon rafflesia, KY513288 (Zhang, Cong, Shen, et al. Citation2017); Tagiades vajuna, KX865091 (Liu et al. Citation2017); Erynnis montanus, KC659955 (Wang et al. Citation2014); Celaenorrhinus maculosa, KF543077 (Wang et al. Citation2015); Daimio tethys, KJ813807 (Zuo et al. Citation2016); Ctenoptilum vasava, JF713818 (Hao et al. Citation2012); Achalarus lyciades, KX249739 (unpublished); Lobocla bifasciatus, KJ629166 (Kim et al. Citation2014); Heteropterus morpheus, KF881050 (unpublished); Carterocephalus silvicola, KJ629163 (Kim et al. Citation2014); Polytremis jigongi, KP765762 (unpublished); Parnara guttata, JX101619 (unpublished); Potanthus flavus, KJ629167 (Kim et al. Citation2014); Lerema accius, KT598278 (Cong and Grishin Citation2016); Ochlodes venata, HM243593 (unpublished); Polytremis nascens, KM981865 (Jiang et al. Citation2016); Choaspes benjaminii, JX101620 (unpublished); Hasora anura, KR189008 (unpublished); Hasora vitta, KR076553 (unpublished); Burara striata, KY524446 (Zhang, Cong, Fan, et al. Citation2017); Megathymus yuccae, KY630500 (Zhang, Cong, Fan, et al. Citation2017); M. streckeri, KY630501 (Zhang, Cong, Fan, et al. Citation2017); M. ursus, KY630502 (Zhang, Cong, Fan, et al. Citation2017); M. cofaqui, KY630503 (Zhang, Cong, Fan, et al. Citation2017); Agathymus mariae, KY630504 (Zhang, Cong, Fan, et al. Citation2017); M. beulahae, KY630505 (Zhang, Cong, Fan, et al. Citation2017); Parnassius bremeri FJ871125 (Kim et al. Citation2009); and Papilio maackii KC433408 (Dong et al. Citation2013).