Figures & data
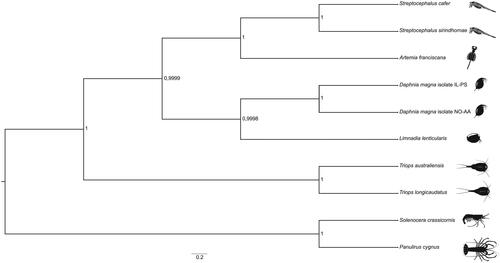
Figure 1. A Bayesian phylogenetic tree constructed in BEAST2 using mitogenome sequences of Streptocephalus cafer (NCBI accession number MN720104) and nine other crustacean species: Streptocephalus sirindhornae (NC_026704.1), Daphnia magna isolate IL-PS (MH683649.1), Daphnia magna isolate NO-AA(MH683655.1), Artemia franciscana (X69067.1), Limnadia lenticularis (NC_039394.1), Panulirus cygnus (KT696496.1), Solenocera crassicornis (KU899137.1), Triops australiensis (LK391946.1) and Triops longicaudatus (AY639934.1). The numbers next to each node represent posterior probability and the scale bar shows the scaled substitution rate.