Figures & data
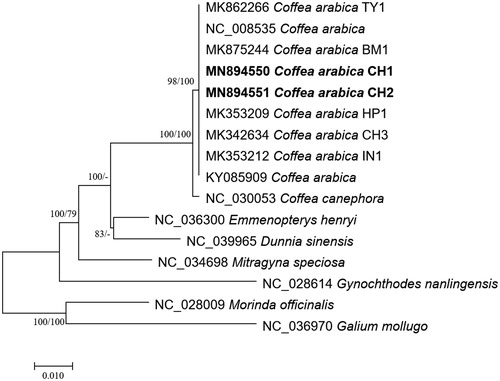
Figure 1 Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of six Coffea and four Rubiaceae complete chloroplast genomes: nine Coffea arabica (MN894550 and MN894551 in this study, MK875244, MK862266, MK353212, NC_008535, KY085909, MK342634, and MK353209), Coffea canephora (NC_030053), Mitragyna speciosa (NC_034698), Dunnia sinensis (NC_039965), Emmenopterys henryi (NC_036300), and Gynochthodes nanlingensis (NC_028614). Phylogenetic tree was drawn based on maximum likelihood tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.