Figures & data
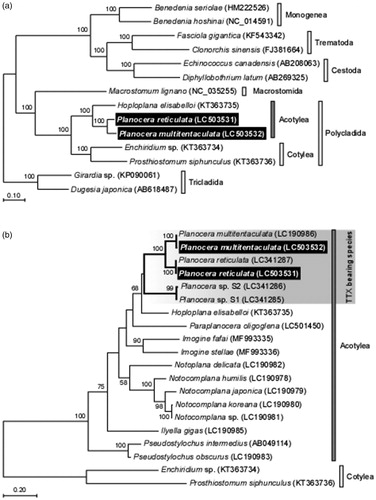
Figure 1. Phylogenetic relationship of two planocerid flatworms related Platyhelminthes species inferred from the 12 protein-coding genes on mitochondrial genome sequence (a) and COI gene (b). The phylogenetic tree was generated by maximum likelihood analysis under the General Time Reversible model. Numbers at branches denote the bootstrap percentages from 1000 replicates. Only bootstrap values exceeding 50% are presented. LC503531 and LC503532 in parentheses indicate the accession numbers deposited in DDBJ/EMBL/GenBank databases in this study and the accession numbers for reference sequences are shown in parentheses. Data used for the analysis were obtained from Planocera reticulata (LC341287), Planocera multitentaculata (LC190986), Hoploplana elisabelloi (KT363735), Planocera sp. S1 (LC341285), Planocera sp. S2 (LC341286), Paraplanocera oligoglena (LC501450), Imogine fafai (MF993335), Imogine stellae (MF993336), Ilyella gigas (LC190985), Notoplana delicata (LC190982), Notocomplana humilis (LC190978), Notocomplana japonica (LC190979), Notocomplana sp. (LC190981), Notocomplana koreana (LC190980), Pseudostylochus obscurus (LC190983), Pseudostylochus intermedius (AB049114), Enchiridium sp. (KT363734), Prosthiostomum siphunculus (KT363736), Macrostomum lignano (NC_035255), Girardia sp. (KP090061), Dugesia japonica (AB184487), Benedenia seriolae (HM222526), Benedenia hoshinai (NC_014591), Fasciola gigantica (KF543342), Clonorchis sinensis (FJ381664), Echinococcus canadensis (AB208063) and Diphyllobothrium latum (AB269325). The scale indicates number of nucleotide substitutions per site.