Figures & data
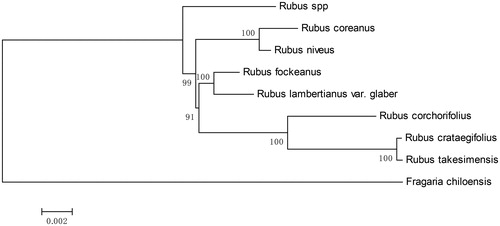
Figure 1. Maximum-likelihood (ML) tree based on conserved collinear blocks of the complete chloroplast genome of Rubus species, using Fragaria chiloensis as an outgroup. The numbers on the node are the fast bootstrap value based on 10,000 replicates. The bar indicates 0.002 mutations per site. Accession No. for the sequences used were listed as: Rubus spp. MH992399, Rubus coreanus MH992398, Rubus niveus KY419961, Rubus fockeanus KY420018, Rubus corchorifolius KY419958, Rubus crataegifolius MG189543, Rubus takesimensis NC_037991, Fragaria chiloensis NC_019601.