Figures & data
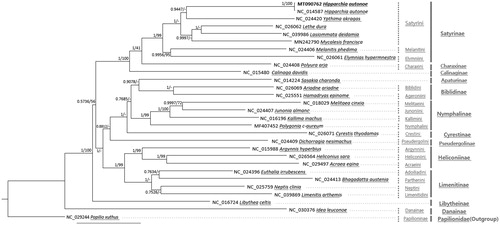
Figure 1. Bayesian inference (1,000,000 generations) and maximum likelihood (1,000 bootstrap repeats) phylogenetic trees based on 28 Nymphalidae mitochondrial genomes: Hipparchia autonoe (MT090762 in this study and NC_014587), Lethe dura (NC_026062), Ypthima akragas (NC_024420), Lasiommata deidamia (NC_039986), Mycalesis francisca (MN242790), Melanitis phedima (NC_024406), Elymnias hypermnestra (NC_026061), Polyura arja (NC_024408), Calinaga davidis (NC_015480), Sasakia charonda (NC_014224), Ariadne ariadne (NC_026069), Hamadryas epinome (NC_025551), Melitaea cinxia (NC_018029), Junonia almana (NC_024407), Kallima inachus (NC_016196), Polygonia c-aureum (MF407452), Cyrestis thyodamas (NC_026071), Dichorragia nesimachus (NC_024409), Argynnis hyperbium (NC_015988), Heliconius sara (NC_026564), Acraea egina (NC_029497), Euthalia irrubescens (NC_024396), Bhagadatta austenia (NC_024413), Neptis clinia (NC_025759), Limenitis arthemis (NC_039869), Libythea celtis (NC_016724), Idea leuconoe (NC_030376), and one Papilionidae species, Papilio xuthus (NC_029244) as an outgroup. Phylogenetic tree was drawn based on Bayesian inference tree. The numbers above branches indicate posterior probability of Bayesian inference tree and bootstrap support value of maximum likelihood phylogenetic tree, respectively. Tribe names are displayed as light gray color and subfamily names were written as dark gray color.