Figures & data
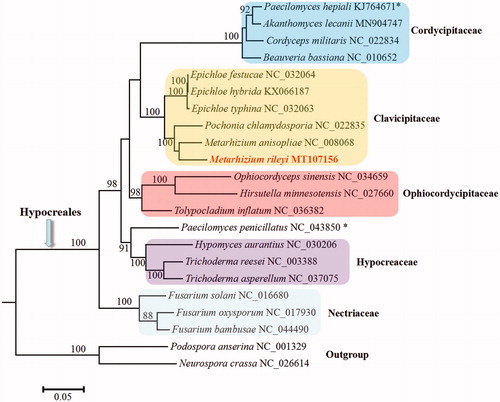
Figure 1. Phylogenetic analysis of Hypocreales species based on mitochondrial nucleotide sequences. We used all species of Clavicipitaceae and representative species of all other families with available mitogenomes in Hypocreales. Two Sordariales species (Podospora anserine and Neurospora crassa) were used as outgroups. The whole mitogenome sequences (or exonic sequences in cases with alignment difficulties) of these species were aligned and trimmed using the HomBlocks pipeline (Bi et al. Citation2018), resulting in an alignment of 7193 characters. Phylogenetic reconstruction was performed using the maximum likelihood approach as implemented in RAxML v8.2.12 (Stamatakis Citation2014). Support values were given for nodes that received bootstrap values ≥ 70%. GenBank accession numbers followed after fungal taxon names. The asterisk (*) indicates fungal taxa with controversial taxonomy in different databases.