Figures & data
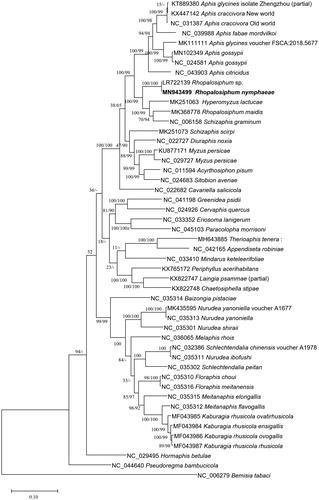
Figure 1. Neighbor joining (10,000 bootstrap repeats) and maximum likelihood (1,000 bootstrap repeats) phylogenetic trees of 48 insect species in the family Aphididae, R. nymphaea (MN943499 and trimmed LR722139 sequence), Rhopalosiphum maidis (MK368778), two Aphis gossypii (MN102349 and NC_024581), Myzus persicae (NC_029727 and KU877171), Acyrthosiphon pisum (NC_011594), Sitobion avenae (NC_024683), Diuraphis noxia (NC_022727), Cavariella salicicola (NC_022682), Schizaphis graminum (NC_006158), Aphis fabae mordvilkoi (NC_039988), Aphis craccivora (NC_031387 and KX447142), Aphis citricidus (NC_043903), Aphis glycines (KT889380 and MK111111), Chaetosiphella stipae (KX822748), Therioaphis tenera (MH643885), Pseudoregma bambucicola (NC_044640), Schizaphis scirpi (MK251073), Hyperomyzus lactucae (MK251063), Hormaphis betula (NC_029495), Cervaphis quercus (NC_024926), Greenidea psidii (NC_041198), Eriosoma lanigerum (NC_033352), Paracolopha morrisoni (NC_045103), Periphyllus acerihabitans (KX765172), Laingia psammae (KX822747), Baizongia pistaciae (NC_035314), Nurudea yanoniella (NC_035313 and MK435595), Nurudea shiraii (NC_035301), Nurudea ibofushi (NC_035311), Schlechtendalia chinensis (NC_032386), Schlechtendalia peitan (NC_035302), Melaphis rhois (NC_036065), Schlechtendalia elongallis (NC_035315), Nurudea choui (NC_035310), Nurudea meitanensis (NC_035316), Schlechtendalia flavogallis (NC_035312), Kaburagia rhusicola ovatirhusicola (MF043985), Kaburagia rhusicola ensigallis (MF043984), Kaburagia rhusicola ovagallis (MF043986), Kaburagia rhusicola rhusicola (MF043987), Mindarus keteleerifoliae (NC_033410), Appendiseta robiniae (NC_042165), and Bemisia tabaci (NC_006279) as an outgroup. Phylogenetic tree was drawn based on neighbor joining tree. The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.