Figures & data
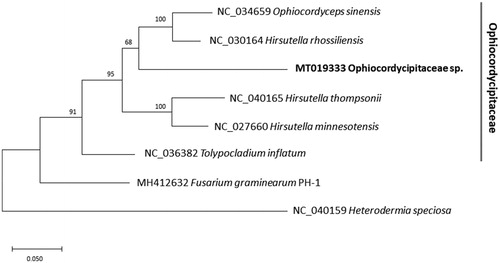
Figure 1. Maximum likelihood (1,000 bootstrap repeats) phylogenetic tree of 12 conserved genes, ATP6, ATP8, COB, COX1, COX2, COX3, NAD1, NAD3, NAD4, NAD4L, NAD5, and NAD6, originated from the six mitochondrial genomes of Ophiocordycipitaceae and two outgroup species: Ophiocordycipitaceae sp., Ophiocordyceps sinensis, Hirsutella rhossiliensis, Hirsutella thompsonii, Hirsutella minnesotensis, and Tolypocladium inflatum and Fusarium graminearum PH-1and Heterodermia speciose as two outgroup species. Gray line indicates family of six fungal species used in the tree. The numbers above branches indicate bootstrap support values of a maximum likelihood phylogenetic tree.