Figures & data
Table 1. Sampling details of Pakistani ungulate species used in this study, including accession numbers, source, and conservation status.
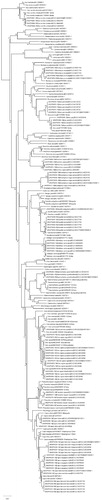
Figure 1. Neighbor-Joining phylogenetic tree of ungulates using COI DNA barcodes. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of base differences per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 5). The analysis involved 188 nucleotide sequences. All positions containing gaps and missing data were eliminated.