Figures & data
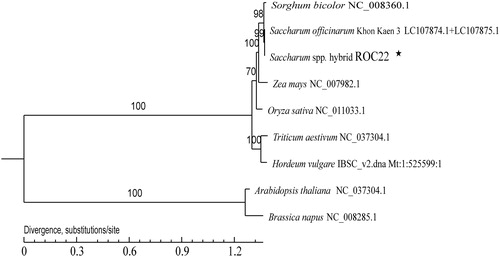
Figure 1. Maximum likelihood tree based on the complete mitochondrial genome sequences of nine species. The numbers on the branches are bootstrap values. The number at each node is the bootstrap probability. The number after the species name is the GenBank accession number. Asterisk, the genome sequence in this study.
Data availability
The data that support the findings of this study are available. Raw data sequences of Saccharum spp. hybrid ROC22 mitchondrial genome are available in NCBI SRA with the accession number of SRR11358605 (BioProject: PRJNA63602) at the URL (https://dataview.ncbi.nlm.nih.gov/objects?linked_to_id=SRR11358605&archive=biosample). The mitogenome of seven species as following are available in the public domain NCBI (Oryza sativa, Zea mays, Sorghum bicolor, Triticum aestivum, Saccharum officinarum Khon Kaen 3, Arabidopisis thaliana, and Brassica napus; the URL https://www.ncbi.nlm.nih.gov/) and the mitogenome of Hordeum vulgare is available in ensembl (http://asia.ensembl.org/index.html).
