Figures & data
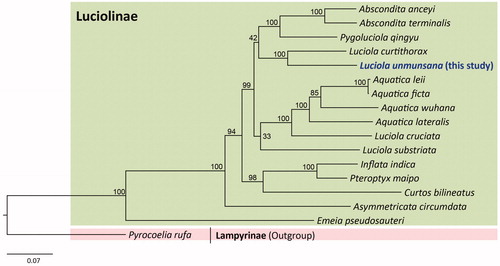
Figure 1. Phylogenetic tree for the subfamily Luciolinae. The maximum likelihood (ML) method was applied using randomized accelerated maximum likelihood (RAxML) ver. 8.0.24 (Stamatakis Citation2014), which was incorporated into the cyberinfrastructure for phylogenetic research (CIPRES) Portal ver. 3.1 (Miller et al. Citation2010). A six optimal partitioning scheme and substitution model (GTR + Gamma + I) were determined using PartitionFinder 2 with the Greedy algorithm (Lanfear et al. Citation2012, Citation2014, Citation2016). Phylogenetic trees were visualized using FigTree ver. 1.42 (http://tree.bio.ed.ac.uk/software/figtree/). The numbers at each node represent bootstrap percentages of 1,000 pseudoreplicates by ML analysis. The scale bar indicates the number of substitutions per site. Lampyrinae (Pyrocoelia rufa, MH352481; Bae et al. Citation2004) is used as an outgroup. GenBank accession numbers are as follows: Abscondita anceyi, MH020192 (Hu and Fu Citation2018b); Abscondita terminalis, MK292092 (Chen et al. Citation2019); Pygoluciola qingyu, MK292093 (Chen et al. Citation2019); Aquatica leii, KF667531 (Jiao et al. Citation2015); Aquatica ficta, KX758085 (Wang et al. Citation2017); Aquatica wuhana, KX758086 (Wang et al. Citation2017); Luciola cruciata, AB849456 (Maeda et al. Citation2017a); Aquatica lateralis, LC306678 (Maeda et al. Citation2017b); Luciola curtithorax, MG770613 (Hu and Fu Citation2018a); Luciola substriata, KP313820 (Mu et al. Citation2016); Inflata indica, MH427718 (Sriboonlert and Wonnapinij Citation2019); Pteroptyx maipo, MF686051 (Fan and Fu Citation2017); Curtos bilineatus, MK292114 (Chen et al. Citation2019); Asymmetricata circumdata, KX229747 (Luan and Fu Citation2016); and Emeia pseudosauteri, MK292112 (Chen et al. Citation2019).
Data availability statement
The data that support the findings of this study are openly available in Mendeley Data at http://dx.doi.org/10.17632/rh3cd4ztx7.1
