Figures & data
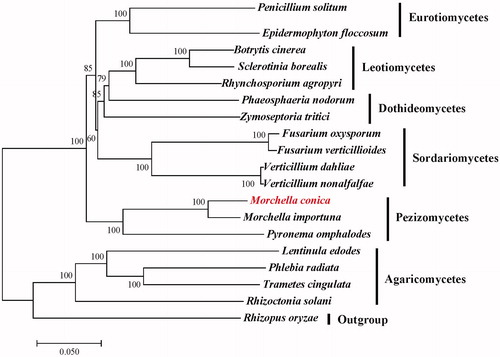
Figure 1. Maximum-likelihood (ML) phylogenetic tree of M. conica and related fungal species. The ML-tree is based on 14 conserved core mitochondrial proteins. Bootstraps values (1,000 replicates) are shown at the nodes. All the sequences are currently available in the GenBank database: Botrytis cinerea (KC832409), Epidermophyton floccosum (NC_007394), Fusarium oxysporum (NC_017930), F. verticillioides (NC_016687), Lentinula edodes (NC_018365), M. importuna (NC_045397), Zymoseptoria tritici (NC_010222), Penicillium solitum (NC_016187), Phaeosphaeria nodorum (NC_009746), Phlebia radiata (NC_020148), Pyronema omphalodes (NC_029745), Rhizoctonia solani (NC_021436), Rhynchosporium agropyri (NC_023125), Sclerotinia borealis (NC_025200), Trametes cingulata (NC_013933), Verticillium dahliae (NC_008248) and V. nonalfalfae (NC_029238). Rhizopus oryzae (NC_006836) was served as an outgroup.
Data availability statement
The data that support the findings of this study are openly available in the Genome Warehouse Database at https://bigd.big.ac.cn/gwh, accession number GWHANVR01000000.
