Figures & data
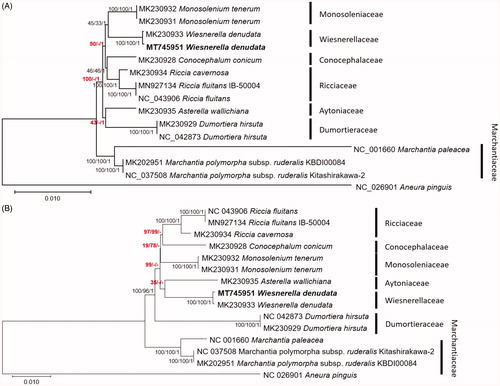
Figure 1. Neighbor-joining (bootstrap repeat is 10,000), maximum likelihood (bootstrap repeat is 1000), and Bayesian Inference (Number of generations is 1,100,000) phylogenetic trees of (A) concatenation of alignment of conserved 37 genes (B) 15 complete mitochondrial genomes: Wiesnerella denudata (MT745951 in this study and MK230933; Dong et al. Citation2019), Monosolenium tenerum (MK230931 and MK230932; Dong et al. Citation2019), Conocephalum conicum (MK230928; Dong et al. Citation2019), Riccia cavernosa (MK230934; Dong et al. Citation2019), Riccia fluitans (NC_043906 and MN927134; Min et al. Citation2020), Asterella wallichiana (MK230935; Dong et al. Citation2019), Dumortiera hirsuta (NC_042873 and MK230929; Dong et al. Citation2019; Kwon et al. Citation2019a), Marchantia polymorpha subsp. ruderalis (NC_037508 and MK202951; Bowman et al. Citation2017; Kwon et al. Citation2019b), Marchantia paleacea (NC_001660; Oda et al. Citation1992), and Aneura pinguis (NC_026901; Myszczyński et al. Citation2017) as an outgroup. Gray bars with labels indicate specific classes. The numbers above branches indicate bootstrap support values of maximum likelihood, neighbor-joining, and Bayesian Inference phylogenetic trees, respectively. Phylogenetic trees were drawn based on maximum likelihood tree. Red-colored supportive values indicate incongruent among three trees.
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MT745951 in NCBI GenBank.
