Figures & data
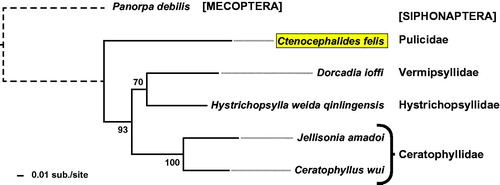
Figure 1. Phylogeny estimation based on five siphonapteran proteomes. Protein sequences (n = 12: ATP synthase F0 subunits 6 & 8, cytochrome b, cytochrome c oxidase subunits I & III, and NADH dehydrogenase subunits 1-6 & 4 L) from 6 sequenced siphonapteran mitogenomes (Ctenocephalides felis, Jellisonia amadoi, Ceratophyllus wui, Dorcadia ioffi, and Hystrichopsylla weida qinlingensis) plus Panorpa debilis (Mecopteran) were independently aligned with MUSCLE v3.8.31 using default parameters. Aligned sequences were concatenated into a single data set (3,505 positions) for phylogeny estimation using RAxML v8.2.4, under the gamma model of rate heterogeneity, WAG model of substitution, and estimation of the proportion of invariant sites. Branch support was assessed with 500 pseudo-replications. Final ML optimization likelihood was -26546.055122.
Data availability statement
The data for this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nucleotide/ under accession number MT594468.
