Figures & data
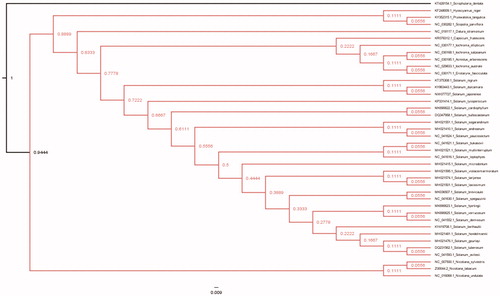
Figure 1. Maximum-Likelihood (bootstrap repeat is 100) phylogenetic trees of S. japonense and related species of Solanaceae based on the whole cp genomes. Chloroplast genomes: GenBank accession numbers: Solanum japonense (MW077727; this study), Solanum gourlayi (MH021476.1), Solanum avilesii (NC_041593.1), Solanum hondelmamii (MH021481.1), Solanum hondelmamii (NC_041608.1), Solanum berthaultii (KY419708.1), Solanum tuberosum (DQ231562.1), Solanum violaceimarmoratum (MH021595.1), Solanum laxissimum (MH021501.1), Solanum tarijense (MH021574.1), Solanum demissum (NC_041552.1), Solanum hjertingii (MK690623.1), Solanum verrucosum (MK690625.1), Solamum microdontum (MH021415.1), Solanum spegazzinii (NC_041630.1), Solanum brevicaule (MK036507.1), Solanum leptophyes (NC_041616.1), Solanum bukasovii (NC_041621.1), Solanum multiinterruptum (MH021521.1), Solanum sogarandinum (MH021551.1), Solanum paucissectum (NC_041624.1), Solanum andreanum (MH021410.1), Solanum cardiophyllum(MK690622.1), Solanum bulbocastanum(DQ347958.1), Solanum lycopersicum (KP331414.1), Solanum dulcamara(KY863443.1), Solanum nigrum (KT375308.1), Scrophularia dentate (KT428154), Acnistus arborescens (NC_030185.1), Capsicum frutescens (KR078312.1), Datura stramonium (NC_018117.1), Eriolarynx fasciculate (NC_030171.1), Hyoscyamus niger (KF248009.1), Iochroma australe (NC_029833.1), Iochroma ellipticum (NC_030177.1), Iochroma salpoanum (NC_030168.1), Nicotiana sylvestris (NC_007500.1), Nicotiana tabacum (Z00044.2), Nicotiana undulata (NC_016068.1), Solanum bulbocastanum cultivar PT29 (DQ347958.Q), Scopolia parviflora (NC_030282.1).
Data availability statement
Solanum_japonense_N cp genome data was submitted to Genbank ( accession number: MW077727).
