Figures & data
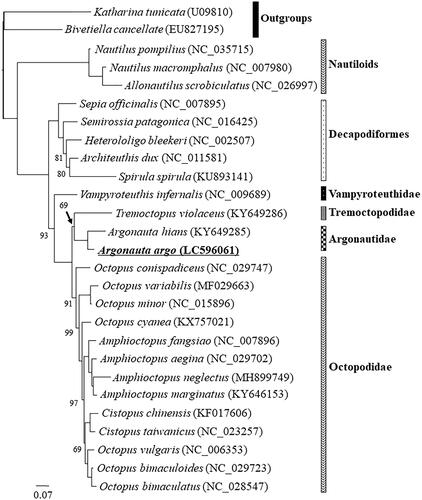
Figure 1. The maximum likelihood (ML) phylogenetic tree showing the position of Argonauta argo. The support values of nodes with 100% bootstrap supports are not shown. Phylogenetic analyses were conducted on a data matrix (10,428 positions) including all concatenated nucleotide sequences of the mitogenomes except the third codon positions of the protein-coding genes. Gene sequences were aligned individually using the online version of MAFFT under default settings (Katoh and Standley Citation2013). Aligned sequences were individually edited using the online version of GBlocks using the least stringent settings (Castresana Citation2000). A partitioned ML analysis (three partitions: protein-coding, rRNA, tRNA) were performed using the RAxML-GUI ver. 1-5b1 (Silvestro and Michalak Citation2012; Stamatakis Citation2014), with the GTR + Γ nucleotide substitution model (Yang Citation1994) 1000 rapid bootstrap replications.
Data availability statement
The full mitochondrial genome sequence reported in this study is registered in and openly available from the National Center for Biotechnology Information (NCBI) Genbank database (Accession No.: LC596061; https://www.ncbi.nlm.nih.gov/nuccore/LC596061.1).
The shell (eggcase) of the specimen was deposited at The University Museum of The University of Tokyo, Japan (Takenori Sasaki; [email protected]) under the voucher number UMUT-RM33391.
