Figures & data
Figure 1. Sampling sites of 21 Culex tritaeniorhynchus geographic populations in the mainland of China.
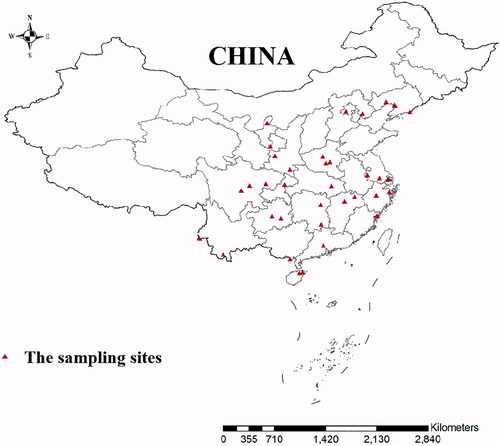
Table 1. Genetic diversity of 21 geographic populations based on COI barcode.
Figure 2. Neighbor-joining tree based on COI barcodes region of C. tritaeniorhynchus. Bootstrap support (1,000 replicates) of nodes from NJ tree and ML tree are indicated above and below the branches, respectively. Only nodes with BS > 70% are labeled.
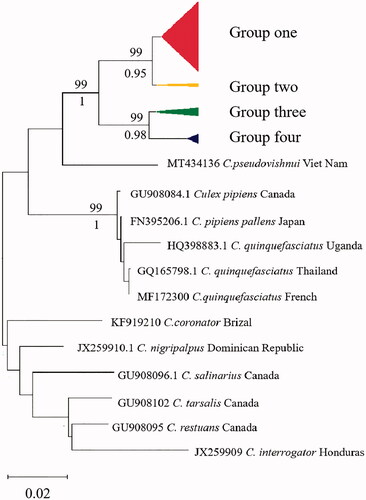
Figure 3. Haplotype network based on COI barcodes of Culex tritaeniorhynchus. Node sizes are proportional to haplotype frequencies. Lines linking the nodes are proportional to the mutation steps.
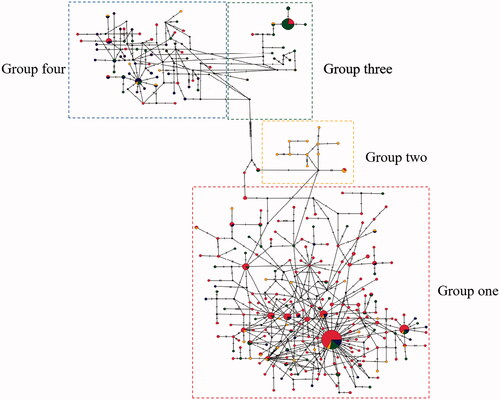
Table 2. AMOVA analysis of C. tritaeniorhynchus populations based on COI gene fragments.
Data availability statement
The data that support the findings of this study are available in the National Center for Biotechnology Information (NCBI) at [https://www.ncbi.nlm.nih.gov/], reference number [MW488441-MW488925]
