Figures & data
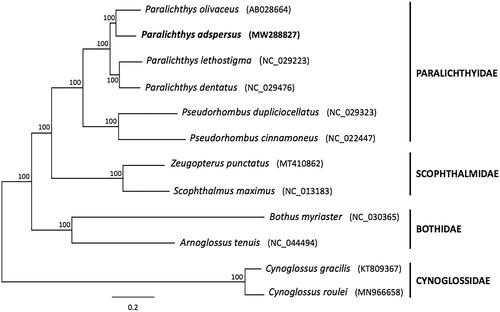
Figure 1. Bayesian phylogenetic tree inferred from 12 concatenated mitochondrial protein-coding genes (ND6 was excluded) of pleuronectiform species. The sequence matrix (10,684 nt) used in the phylogenetic analyses consisted of unambiguously aligned regions of the first, second, and third codon positions (COI and ATPase 8), all other PCGs included only first and second codon positions. The GTR + I+G model was estimated as the best-fit substitution model for genes ATPase6, COII, ND1, ND2, and ND5; HKY + G model for genes ATPase8 and COI; HKY + I+G model for genes COIII, Cytb, and ND4; and GTR + G model for genes ND3 and ND4L. The position of Paralichthys adspersus (whose mitogenome was determined in this study) is shown in bold. Posterior probabilities at correspondent nodes are shown in percentages. GenBank accession numbers for each species are shown in parentheses.
Data availability statement
The data that support the findings of this study are openly available in [GenBank reference number MW288827, https://www.ncbi.nlm.nih.gov/nuccore/MW288827].
