Figures & data
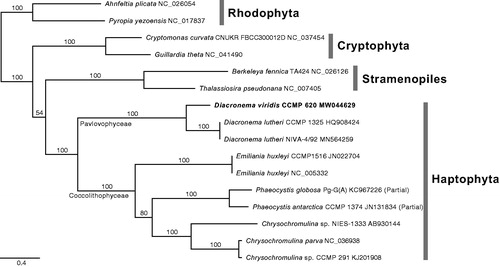
Figure 1. The maximum-likelihood mtDNA phylogeny of haptophytes based on 15 combined CDS data, including 10 haptophytes and six outgroups (Cryptophytes, Rhodophyta, and Stramenopiles). All CDS were aligned with translated amino acids and then concatenated as a total of 13,254 positions without ambiguous regions. The best phylogeny inferred under the GTR with independent heterogeneity (GTR + G) model for total 45 partitions, i.e. three codon positions of each gene. Bootstrap support values were calculated with 1,000 replicates using the same substitution model. The best phylogeny supports sister relationship of Diacronema viridis and D. lutheri with maximum bootstrap value.
Data availability statement
Newly determined mitogenome is available in on-line at https://www.ncbi.nlm.nih.gov under the accession no. MW044629. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA691634, SRP301484, and SAMN17301202, respectively.
