Figures & data
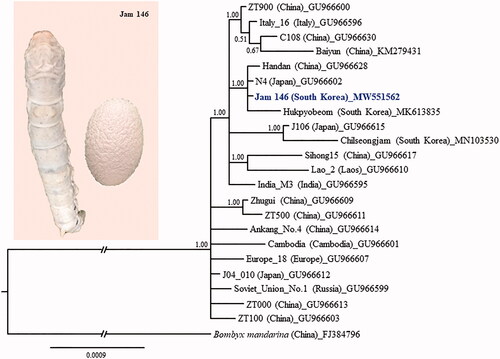
Figure 1. Phylogenetic tree of Bombyx mori. The Bayesian inference (BI) method, based on the concatenated sequences of 13 protein-coding genes and two ribosomal RNAs, was used for phylogenetic analysis. The GenBank accession numbers are provided next to strain name. The numbers at the node specify Bayesian posterior probabilities. The scale bar indicates the number of substitutions per site. The wild silkworm, Bombyx mandarina (Hu et al. Citation2010), was designated as the outgroup based on previous phylogenetic results (Wang et al. Citation2019). Publication information is as follows: Baiyun, Zhang et al. (Citation2016); Jam 146, This study; Hukpyobeom, Kim et al. (Citation2019a); Chilseongjam, Kim et al. (Citation2019b); and the remaining strains, Li et al. (Citation2010). The branch length was truncated to approximately one-third of the original length due to limited space.
Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW551562.1
