Figures & data
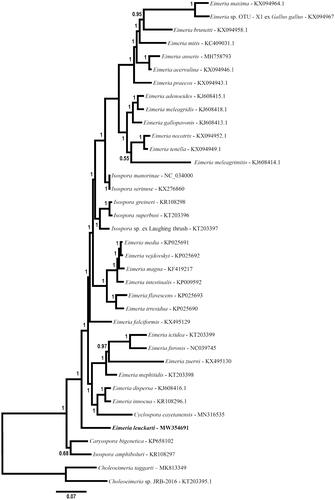
Figure 1. Phylogenetic relationship of the mt genome of Eimeria leuckarti among mitochondrial genomes of various eimeriid parasites inferred by Bayesian analysis of concatenated coding regions (COI, COIII, CytB, rDNA fragments). The resulting alignment was partitioned for Bayesian analysis as follows: CDS partition was analyzed using a codon-based nucleotide substitution model (i.e. Nucmodel = Codon; Code = MetMt) and the rDNA partition was analyzed simultaneously using a simpler general time reversible nucleotide substitution model with allowance for invariant characters and gamma distributed rate variation among sites (i.e. GTR + I + G; Nst = 6; Nucmodel = 4by4). Two Choleoeimeria species were used to root the tree. Horizontal distance is proportional to hypothesized evolutionary change and posterior probability support for each node is indicated.
Data availability statement
The genome sequence data are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under accession no. MW354691, and additional data that support the findings of this study are available at Mendeley Data at http://dx.doi.org/10.17632/j6tjg4sdjf.2.
