Figures & data
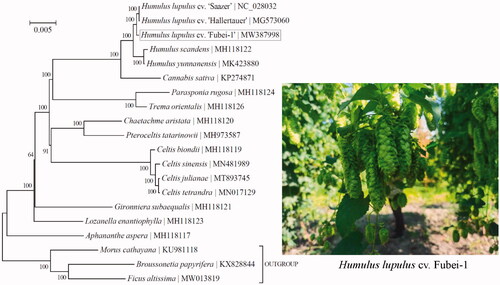
Figure 1. Phylogeny of 17 taxa within the family Cannabaceae based on the Bayesian analysis of chloroplast protein-coding genes. The best-fit nucleotide substitution model is ‘GTR + G+I’. The support values were placed next to the nodes. Three taxa within the family Moraceae (i.e. Broussonetia papyrifera, Ficus altissima, and Morus cathayana) were included as outgroup taxa.
Data availability statement
The data that support the findings of this study are openly available in GenBank from NCBI at https://www.ncbi.nlm.nih.gov under the accession number MW387998.
