Figures & data
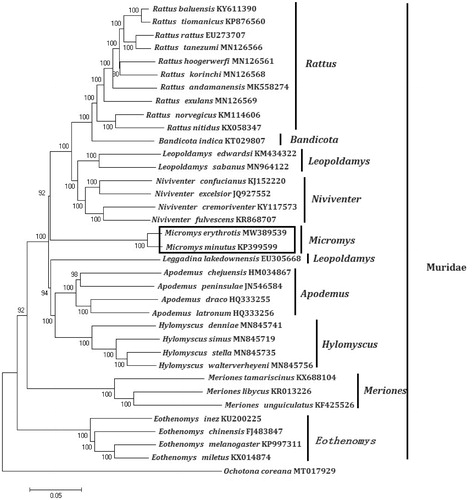
Figure 1. Phylogenetic tree generated using the Maximum Likelihood method based on complete mitochondrial genomes. Rattus andamanensis (MK558274), Rattus nitidus (KX058347), Rattus rattus (EU273707), Rattus norvegicus (KM114606), Rattus tiomanicus (KP876560), Rattus exulans (MN126569), Rattus korinchi (MN126568), Rattus tanezumi (MN126566), Rattus hoogerwerfi (MN126561), Rattus baluensis (KY611390), Niviventer fulvescens (KR868707), Niviventer confucianus (KJ152220), Niviventer excelsior (JQ927552), Niviventer cremoriventer (KY117573), Bandicota indica (KT029807), Hylomyscus denniae (MN845741), Hylomyscus walterverheyeni (MN845756), Hylomyscus stella (MN845735), Hylomyscus simus (MN845719), Leopoldamys edwardsi (KM434322), Leopoldamys sabanus (MN964122), Apodemus peninsulae (JN546584), Apodemus latronum (HQ333256), Apodemus draco (HQ333255), Apodemus chejuensis (HM034867), Micromys minutus (KP399599), Micromys erythrotis (MW389539), Eothenomys miletus (KX014874), Eothenomys melanogaster (KP997311), Eothenomys chinensis (FJ483847), Eothenomys inez (KU200225), Leggadina lakedownensis (EU305668), Meriones tamariscinus (KX688104), Meriones libycus (KR013226), Meriones unguiculatus (KF425526). The out group is Ochotona coreana (MT017929).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/, reference number MW389539.
