Figures & data
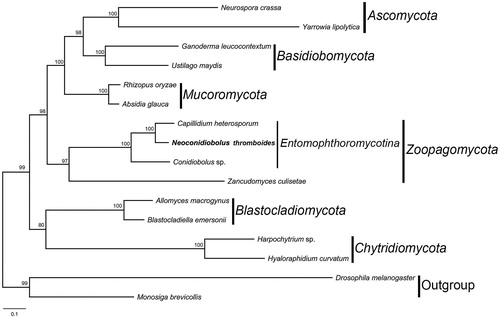
Figure 1. The phylogenetic tree of 14 fungal taxa constructed by maximum likelihood algorithm based on 14 translated mitochondrial proteins. These proteins include oxidase subunits (Cox1, 2, and 3), the apocytochrome b (Cob), ATP synthase subunits (Atp6, Atp8, and Atp9), NADH dehydrogenase subunits (Nad1, 2, 3, 4, 5, 6, and Nad4L). Along with the mitogenome of Neoconidiobolus thromboides, other 13 fungal mitogenomes were used in this phylogenetic analysis: Absidia glauca (NC_036158), Allomyces macrogynus (NC_001715), Blastocladiella emersonii (NC_011360), Capillidium heterosporum (NC_040967), Conidiobolus sp. (MN_640580), Ganoderma leucocontextum (NC_037937), Harpochytrium sp. (NC_004623), Hyaloraphidium curvatum (NC_003048), Neurospora crassa (NC_026614), Rhizopus oryzae (NC_006836), Ustilago maydis (NC_008368), Yarrowia lipolytica (NC_002659), and Zancudomyces culisetae (NC_006837). Besides, Drosophila melanogaster (NC_024511) and Monosiga brevicollis (NC_004309) were served as outgroups. Maximum-likelihood bootstrap values (500 replicates) of each clade are indicated along branches. Scale bar indicates substitutions per site.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, Reference no. MW795364.
