Figures & data
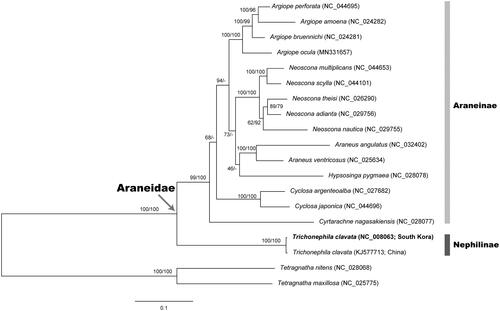
Figure 1. A maximum-likelihood (ML) tree reconstructed with the nucleotide sequence alignment set (without 3rd codon position) of 13 mitochondrial protein-coding genes (PCGs) showing relationships among 19 spider species belonging to the family Araneidae. Two tetragnathid spider species Tetragnatha nitens and Tetragnatha maxillosa are used as outgroups. The ML tree based on amino acid sequence alignment set from 13 PCGs yielded almost same topology with the nucleotide-based ML tree. Nodal supports are inferred from the ultrafast bootstrap method with 1000 bootstrap replicates using the IQ-TREE webserver: bootstrapping values in percent (BP) obtained using the nucleotide data set (left) and amino acid data set (right) on each node.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/NC_008063.1.
