Figures & data
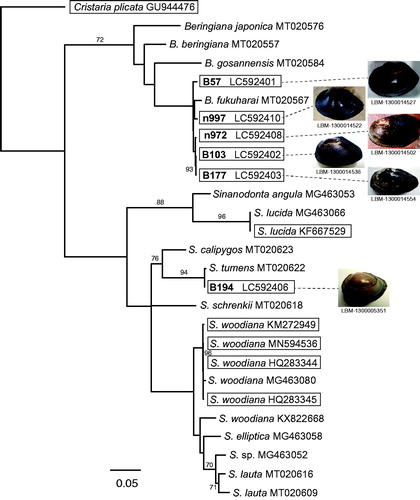
Figure 1. Supermatrix tree of 12 mitogenomes (15,708–16,385 bp) and 15 partial COI sequences (597–657 bp) of female-transmitted (F) mitogenomes of Sinanodonta and Beringiana species (Cristaria plicata used as an outgroup). Bootstrap support (≥70%) is indicated at the nodes. For the previously identified sequences (15 partial sequences and six mitogenomes), accession numbers are given after the species names. For the six mitogenomes sequenced here, the accession numbers are indicated after the specimen IDs used in Mabuchi and Nishida (Citation2020) (the IDs are in bold). The six newly sequenced and six previously published mitogenomes are boxed. The tree backbone was first generated for the 12 mitogenomes by the neighbor-joining (NJ) method using the online version of MAFFT (https://mafft.cbrc.jp/alignment/server/). The obtained NJ tree was then used as a backbone constraint for the supermatrix tree, which was constructed based on the dataset including the 12 mitogenomes and 15 partial sequences, which were first aligned using MAFFT and corrected by eye using Mesquite (version 3.31; http://www.mesquiteproject.org). After deleting the intergenic region, maximum likelihood analysis was performed for the resultant 14,785-bp dataset using RAxML BlackBox (https://embnet.vital-it.ch/raxml-bb/).
Data availability statement
The genome sequence data supporting this study are openly available in NCBI and DDBJ at nucleotide database, https://www.ncbi.nlm.nih.gov/nuccore/LC592401, LC592402, LC592403, LC592408, LC592410, LC592406, Associated BioProjects, https://www.ncbi.nlm.nih.gov/bioproject/PRJDB10947, PRJDB11033, PRJDB11034, PRJDB11035, PRJDB11036, PRJDB11037, BioSample accession numbers at https://www.ncbi.nlm.nih.gov/biosample/SAMD00265731, SAMD00271493, SAMD00271494, SAMD00271495, SAMD00271496, SAMD00271503, and Sequence Read Archives at https://ddbj.nig.ac.jp/DRASearch/submission?acc=DRA011399, DRA011400, DRA011401, DRA011402, DRA011403, DRA011404, DRA011405, DRA011406, DRA011407, DRA011408, DRA011409, DRA011410, DRA012100, DRA012101, DRA012102, DRA012103, DRA012104, DRA012105.
