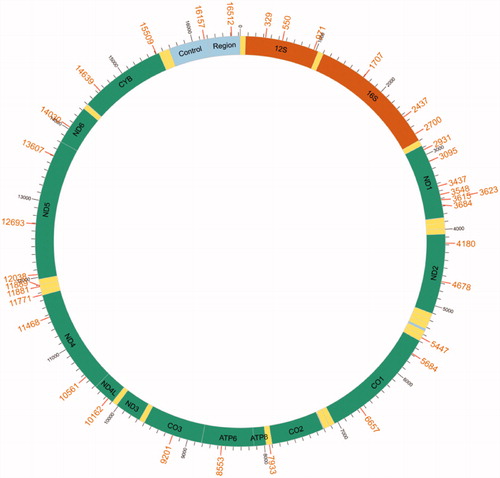
Figures & data
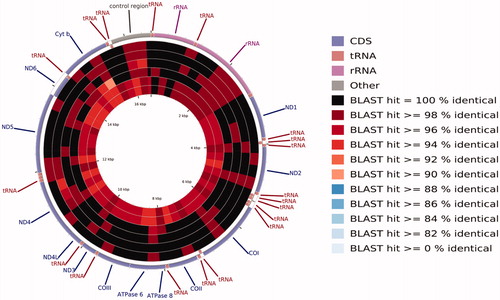
Figure 1. A BLAST-based pie chart of the sequence identity of the mitochondrial genomes of Chanodichthys mongolicus, Ancherythroculter wangi, and Ancherythroculter kurematsui from the different geographical populations. The circle from outside to inside represents Chanodichthys mongolicus MZ032228 from China's Qiantang River, Ancherythroculter wangi MG783573 from China's Nei River, Chanodichthys mongolicus AP009060 from Russia's Black River, Chanodichthys mongolicus KF826087, Chanodichthys mongolicus KC701385 from China's Heilong River, Ancherythroculter kurematsui KU234534 from China's Jialing River, and Ancherythroculter wangi MG575902 from China's Qi River
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ032228. The associated BioProject, SRA, and BioSample numbers are PRJNA732476, SRX10977011, and SAMN19321491, respectively.