Figures & data
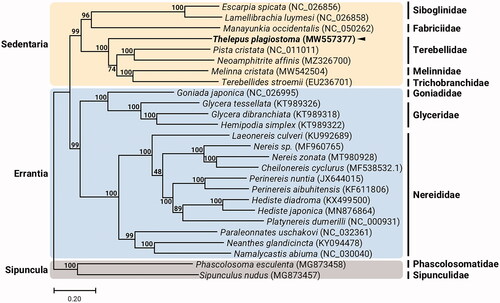
Figure 1. Maximum-likelihood (ML) phylogeny of 7 published mitogenomes from Sedentaria including T. plagiostoma and 17 registered mitogenomes of Errantia species, and two Sipuncula species as an outgroup based on the concatenated nucleotide sequences of protein-coding genes (PCGs). The phylogenetic analysis was performed using the maximum likelihood method, GTR + G + I model with a bootstrap of 1,000 replicates. Numbers on the branches indicate ML bootstrap percentages. DDBJ/EMBL/Genbank accession numbers for published sequences are incorporated. The black triangle means the polychaete analyzed in this study.
Data availability statement
BioProject, BioSample, and SRA accession numbers are https://www.ncbi.nlm.ni h.gov/bioproject/PRJNA699104, https://www.ncbi.nlm.nih.gov/biosample/SAMN17766233, and https://www.ncbi.nlm.nih.gov/sra/?term=SRR15041259, respectively. The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, with an accession number MW557377.
