Figures & data
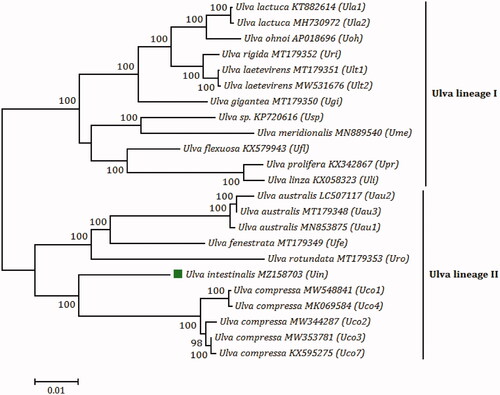
Figure 1. The maximum likelihood (ML) phylogenomic tree was constructed with 1000 bootstrap replicates based on amino acid (aa) sequences of 71 common PCGs from chloroplast genomes of 15 Ulva species, using MEGA 7.0. U. compressa MK069585 (Uco5) and U. compressa MT916929 (Uco6) were absent in this tree, due to their incomplete chloroplast genomes and faulty assembly of some PCGs (rpoA, rpoB, rpoC1, and rpoC2) (Liu and Melton Citation2021). Branch lengths are proportional to the amount of sequence change, which are indicated by the scale bar below the trees.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI under the accession number MZ158703 (https://www.ncbi.nlm.nih.gov/nuccore/MZ158703.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA728799 (https://www.ncbi.nlm.nih.gov/bioproject/728799), SRR14493638 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR14493638), and SAMN19103542 (https://www.ncbi.nlm.nih.gov/biosample/SAMN19103542), respectively.
