Figures & data
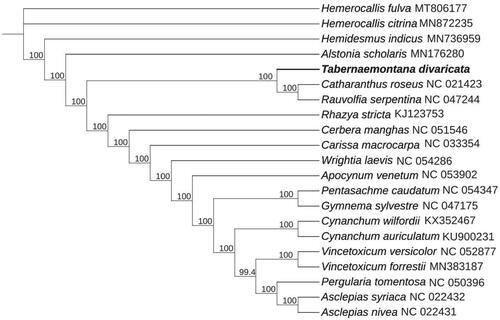
Figure 1. Maximum-likelihood phylogenetic tree for T. divaricata based on nine complete cp genomes of Apocynaceae, 12 complete cp genomes within Asclepiadaceae family, and two species (Hemerocallis citrina and Hemerocallis fulva) from Asphodelaceae as outgroup. The bootstrap values are located on each node and the Genbank accession numbers are shown beside the Latin name of the species.
Data availability statement
The assembled complete cp genome sequence of T. divaricata has been submitted to GenBank of NCBI and is openly available under the accession number: MZ073339 (https://www.ncbi.nlm.nih.gov/nuccore/MZ073339.1/). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA726627, SRR14373633, and SAMN18951226, respectively.
