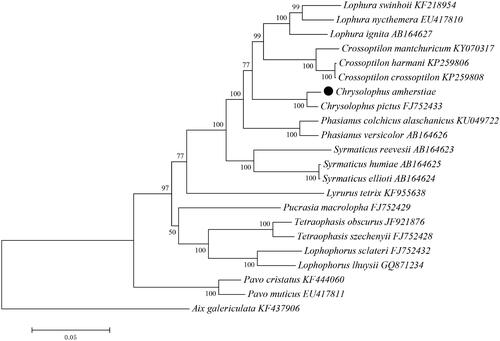
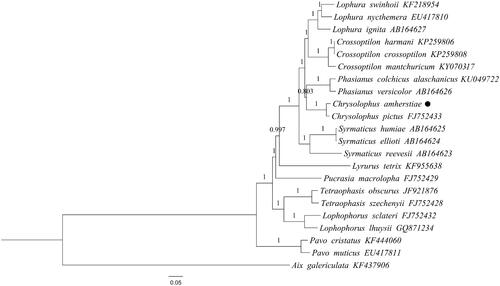
Figures & data
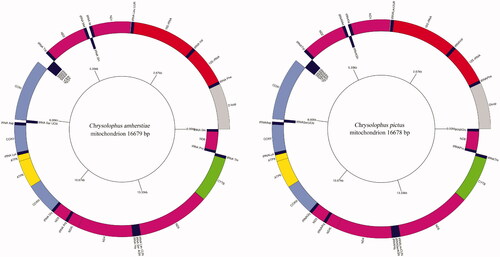
Table 1. Annotation of the mitochondrial genome of Chrysolophus amherstiae (C.a) and Chrysolophus pictus (C.p).
Table 2. Nucleotide composition in PCGs of mitogenomes of Chrysolophus amherstiae (C.a) and Chrysolophus pictus (C.p).
Figure 2. The relative synonymous codon usage (RSCU) in the mitogenomes of C. amherstiae and C. pictus.
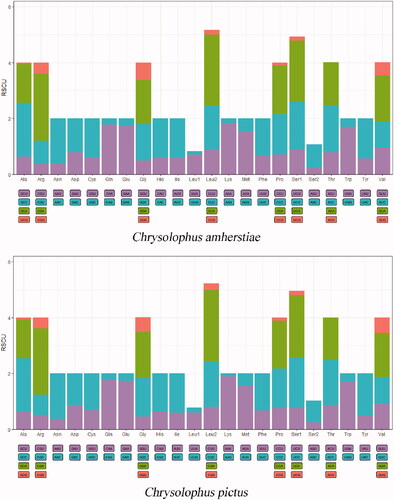
Figure 3. Prognostic map of 12S rRNA secondary structures in C. amherstiae and C. pictus. The different bases are shown in red. I–IV represent structural domains.
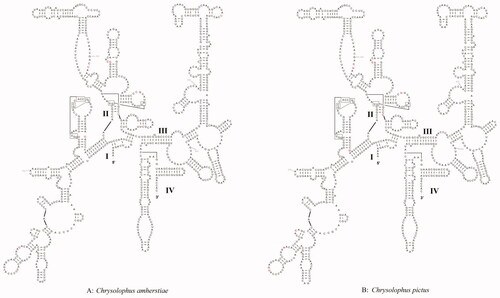
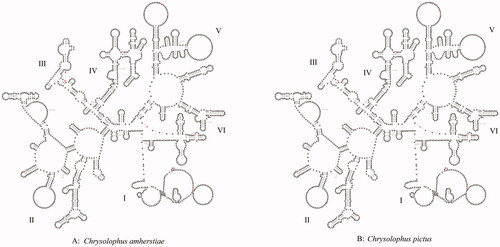
Figure 4. Prognostic map of 16S rRNA secondary structures in C. amherstiae and C. pictus. The different bases are shown in red. The red arrow represents the inserted base. I–VI represent structural domains.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW880933.