Figures & data
Data availability statement
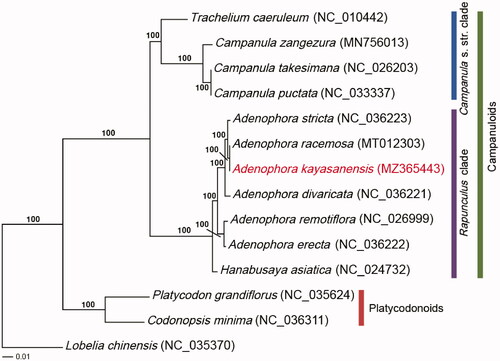
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ365443. The associated BioProject, SRA, and bio-sample numbers are PRJNA736621, SRR14777593, and SAMN19655181, respectively.