Figures & data
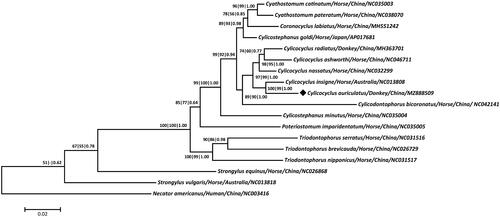
Figure 1. Phylogeny was inferred from maximum parsimony (MP), maximum-likelihood (ML) and Bayesian inference (BI) analyses based on concatenated amino-acid sequences of 12 mt protein-coding genes of C. auriculatus and other related nematodes. Numbers along the branches represent bootstrap values calculated from different analyses in the order: MP/ML/BI; values < 50% are not shown. The scale indicates an estimate of substitutions per site, using the optimized model setting. The solid black diamond represents the species in this study.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, under the accession number MZ888509. The associated BioProject, SRA and Bio-Sample numbers are PRJNA794955, SRR17475793 and SAMN24665689, respectively.
