Figures & data
Figure 1. Specimen of Devario interruptus was collected from the Yingjiang County, Yunnan Province, China. Photograph by Xiao Jiang CHEN on Jun 10, 2021.

Figure 2. Complete mitochondrial genome map of Devario interruptus (GenBank: MZ853154), with 13 protein coding genes, 22 tRNAs, 2 rRNAs, and 2 non-coding regions. Genes encoded on light strand and heavy-strand were shown inner and outside of the gray circle respectively.
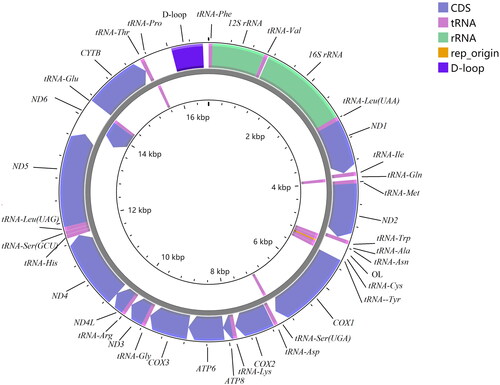
Table 1. Characteristics of the mitochondrial genome of Devario interruptus.
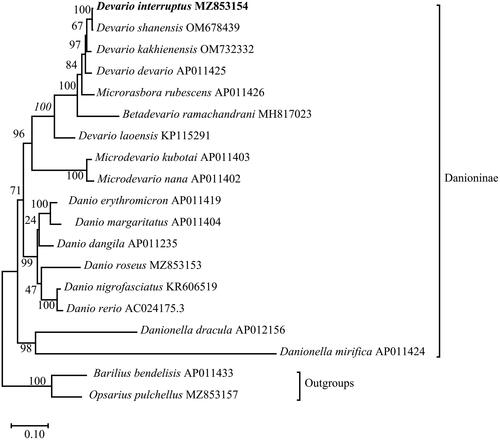
Figure 3. Phylogenetic reconstruction of D. interruptus and other 18 species based on the concatenated mitochondrial protein-coding genes using Maximum-likelihood method. Barilius bendelisis and Opsarius pulchellus were set to be outgroups. Numbers near the nodes indicated bootstrap support values from 1000 replicates.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ853154. The associated "BioProject", "Bio-Sample" and "SRA" numbers are PRJNA769978, SAMN22187307, SRR16301567, respectively.
