Figures & data
Figure 1. Photos of the jack silverside, Atherinopsis californiensis. (A) Jack silverside, commonly also called jacksmelt, belongs to the family of silversides (family Atherinopsidae) and not the taxon of ‘true’ smelts (family Osmeridae). Jack silverside are distinguished from another morphologically similar species – the topsmelt silverside (Atherinops affinis) – by the anterior edge of the anal fin (dotted line) being positioned between the posterior edge of the first dorsal fin and anterior boundary of the second dorsal fin (arrows). (B) Adult jack silverside showing the typical yellow color on the gill operculum, with positioning of the two dorsal fins stretched upright. Photos by Sean Lema.

Figure 2. Circular sketch map of the complete mitogenome of the jack silverside Atherinopsis californiensis. Positions of amino acid coding sequence (CDS) genes, rRNAs, and tRNAs are indicated, as is GC nucleotide composition variation. Visualization generated using the Proksee server (https://proksee.ca/), which utilizes GCView (Stothard and Wishart Citation2005) for circular genome drawing.
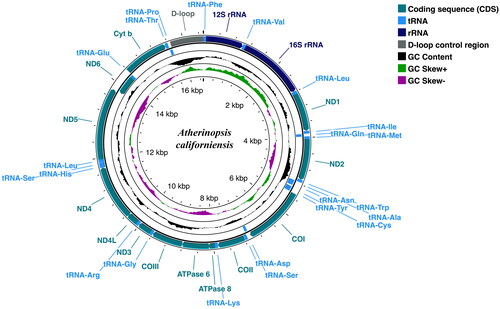
Table 1. Location and arrangement of genes on the mitogenome of A. californiensis.
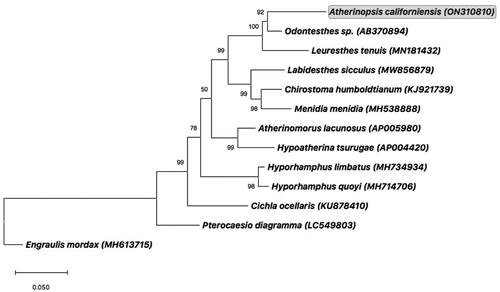
Figure 3. Consensus maximum-likelihood phylogenetic tree of the complete mitogenome of jack silverside Atherinopsis californiensis (GenBank accession no. ON310810; shown in grey box) and other fishes of Order Atheriniformes. GenBank accession numbers are provided in parentheses accompanying each taxon. The tree was rooted using the complete mitogenome from the Northern Anchovy, Engraulis mordax (Lewis and Lema Citation2019). Additional information on previously published complete mitogenomes for the following species used in this phylogenetic analysis can be found as follows: Odontesthes sp. (AB370894) (Setiamarga et al. Citation2008); Leuresthes tenuis (MN181432) (Muñoz et al. Citation2020); Chirostoma humboldtianum (KJ921739) (Barriga-Sosa et al. Citation2016); Menidia menidia (MH538888) (Lou et al. Citation2018); Hypoatherina tsurugae (AP004420) (Miya et al. Citation2003); Hyporhamphus limbatus (MH734934) (Lü et al. Citation2018); Hyporhamphus quoyi (MH714706) (Zhu et al. Citation2018), and Cichla ocellaris (KU878410) (Lin et al. Citation2017).
Data availability
The mitochondrion genome sequence data that support the findings of this study are openly available in GenBank of the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/ under accession no. ON310810. Raw 150PE reads generated by the authors to assemble this mitogenome are available under GenBank BioProject no. PRJNA824876, BioSample no. SAMN27483993, and Run Sequence Read Archive no. SRR18689888.
