Figures & data
Figure 1. A branch of Lophostemon confertus showing the morphology of leaves and fruits. Photographed by Wenhang Su.

Figure 2. The chloroplast genome map of Lophostemon confertus. Genes are shown outside and inside the outer circle are transcribed counterclockwise and clockwise, respectively. GC and AT contents across the chloroplast genome are shown with dark and light shading, respectively, inside the inner circle.
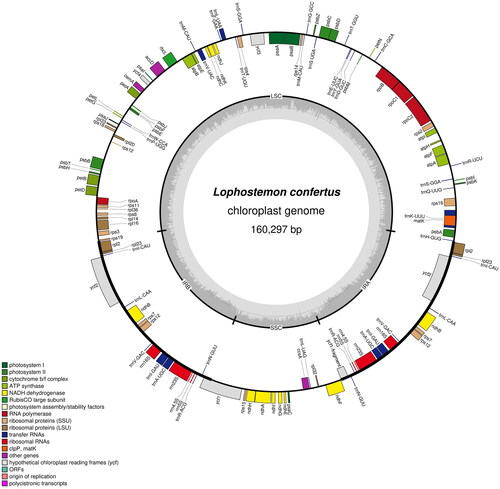
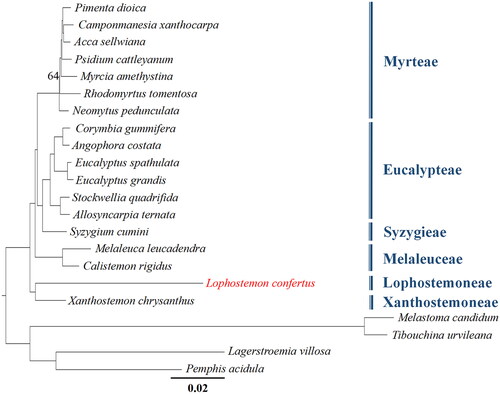
Figure 3. Phylogenetic tree based on the complete chloroplast genome sequences of 18 species from Myrtaceae, with four species from Melastomateae and Lythraceae, used as outgroups. Bootstrap support values of all but one node are 100 (omitted here), and only one node with a bootstrap support value of 64 is shown. The following sequences were used: Pimenta dioica KY085891.1 (unpublished), Camponmanesia xanthocarpa KY392760.1 (unpublished), Acca sellwiana KX289887.1 (Machado et al. Citation2017), Psidium cattleyanum MN095413.1 (Rodrigues et al. Citation2020), Myrcia amethystina MW353255.1 (Lima et al. Citation2021), Rhodomyrtus tomentosa MK044696.1 (Huang et al. Citation2019), Neomytus pedunculata MW214671.1 (Maurin et al. Citation2022), Corymbia gummifera KC180800.1 (Bayly et al. Citation2013), Angophora costata KC180805.1 (Bayly et al. Citation2013), Eucalyptus spathulata KC180793.1 (Bayly et al. Citation2013), Eucalyptus grandis MT700491.1 (unpublished), Stockwellia quadrifida KC180807.1 (Bayly et al. Citation2013), Allosyncarpia ternata KC180806.1 (Bayly et al. Citation2013), Syzygium cumini GQ870669.3 (unpublished), Melaleuca leucadendra MT700493.1 (unpublished), Calistemon rigidus MN794317.1 (Liu et al. Citation2020), Xanthostemon chrysanthus MW837774.1 (unpublished), Melastoma candidum KY745894.1 (Ng et al. Citation2017), Tibouchina urvileana NC043810.1 (Gonçalves et al. Citation2019), Lagerstroemia villosa MK881633.1 (Gu et al. Citation2019), Pemphis acidula MH727532.1 (Jian and Ren Citation2019).
Supplemental Material
Download PNG Image (251.4 KB)Supplemental Material
Download PNG Image (392.3 KB)Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/OM640421 under the accession no. OM640421. And the associated Bioproject, SRA, Bio-sample numbers are PRJNA804362, SRR17931549 and SAMN25731117, respectively
