Figures & data
Figure 1. The picture of the collected sample of S. bulbiferum. The picture is self-taken by Zijie Deng at Changde Baima Lake Cultural Park, Changde, Hunan province, China (29°03′6.55ʺN, 111°39′59.54ʺE; 33 m). In the picture, S. bulbiferum is the wide-spread plant with small leaves, which has a fibrous root, 7–22 cm long stems, white globular bulbils that are viviparous, and a 3-branched and numerous-flowered cyme.

Figure 2. Gene map of the S. bulbiferum chloroplast genome. From the center outward, the first track indicates the dispersed repeats; The second track shows the long tandem repeats as short blue bars; The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors; The fourth track shows small single-copy (SSC), inverted repeat (Ira and Irb), and large single-copy (LSC) regions. The GC content along the genome is plotted on the fifth track; The genes are shown on the sixth track.
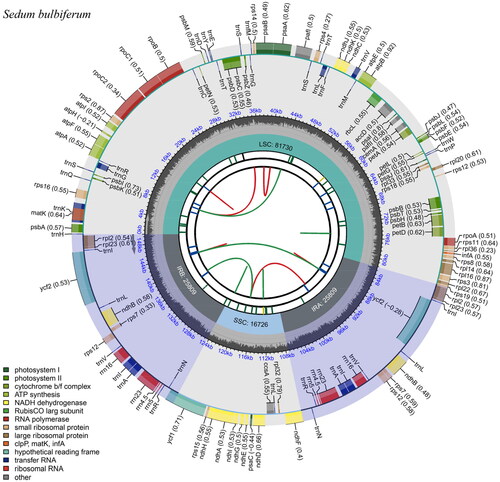
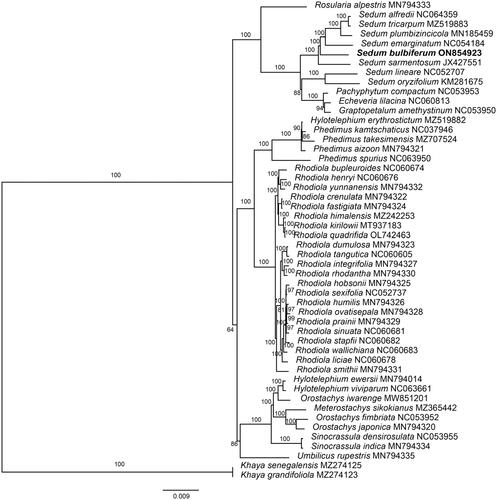
Figure 3. Maximum-likelihood (ML) tree of S. bulbiferum and 49 relative species was reconstructed using the IQ-Tree based on 55 protein-coding gene sequences shared by all genomes under the TVM + I + F model for 5000 ultrafast bootstraps, as well as the Shimodaira–Hasegawa–like approximate likelihood-ratio test. Bootstrap values are shown next to the nodes. The following sequences were used: Rosularia alpestris MN794333 (Zhao et al. Citation2020), Sedum tricarpum MZ519883 (Chen et al., Citation2022), Sedum plumbizincicola MN185459 (Ding et al. Citation2019), Sedum lineare NC052707 (Tang et al. Citation2021), Sedum oryzifolium KM281675 (Li and Chen Citation2020), Echeveria lilacina NC060813 (Nah et al. Citation2022), Hylotelephium erythrostictum MZ519882 (Chen et al., Citation2022), Phedimus kamtschaticus NC037946 (Seo and Kim Citation2018), Phedimus takesimensis MZ707524 (Seo et al. Citation2020), Phedimus aizoon MN794321 (Zhao et al. Citation2020), Rhodiola bupleuroides NC060674 (Zhao et al. Citation2021), Rhodiola henryi NC060676 (Zhao et al. Citation2021), Rhodiola yunnanensis MN794332 (Zhao et al. Citation2021), Rhodiola crenulata MN794322 (Zhao et al. Citation2020), Rhodiola fastigiata MN794324 (Zhao et al. Citation2020), Rhodiola kirilowii MT937183 (Zhang and Liu Citation2021), Rhodiola quadrifida OL742463 (Zhao et al. Citation2022), Rhodiola dumulosa MN794323 (Zhao et al. Citation2020), Rhodiola tangutica NC060605 (Lakshmanan et al. Citation2011), Rhodiola integrifolia MN794327 (Zhao et al. Citation2020), Rhodiola rhodantha MN794330 (Zhao et al. Citation2020), Rhodiola hobsonii MN794325 (Zhao et al. Citation2020), Rhodiola humilis MN794326 (Zhao et al. Citation2020), Rhodiola ovatisepala MN794328 (Zhao et al. Citation2020), Rhodiola prainii MN794329 (Zhao et al. Citation2020), Rhodiola sinuata NC060681 (Zhao et al. Citation2021), Rhodiola stapfii NC060682 (Zhao et al. Citation2021), Rhodiola wallichiana NC060683 (Zhao et al. Citation2021), Rhodiola liciae NC060678 (Zhao et al. Citation2021), Rhodiola smithii MN794331 (Zhao et al. Citation2020), Hylotelephium ewersii MN794014 (Zhao et al. Citation2020), Meterostachys sikokianus MZ365442 (Kang et al. Citation2022), Orostachys japonica MN794320 (Zhao et al. Citation2020), Sinocrassula indica MN794334 (Zhao et al. Citation2020), Umbilicus rupestris MN794335 (Zhao et al. Citation2020), Khaya grandifoliola MZ274123 (Mascarello et al. Citation2021), Khaya grandifoliola MZ274125 (Mascarello et al. Citation2021).
Supplemental Material
Download MS Word (334.3 KB)Data availability statement
The complete chloroplast genome sequence of Sedum bulbiferum has been deposited in the GenBank database under the accession number ON854923 (https://www.ncbi.nlm.nih.gov/nuccore/ON854923). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA867513, SRR20981864, and SAMN30203515, respectively.
