Figures & data
Figure 1. Plant morphological characteristics of Crataegus scabrifolia. (A) Leaves of this species ovate-lanceolate to ovate-elliptic. (B) The flowers of C. scabrifolia are corymbos or compound corymbos. The photos of C. scabrifolia were taken by the authors in Luoping Mountain, Eryuan County, Yunnan Province, China (coordinates: 99°52′19.15″E, 25°59′53.34″N).

Figure 2. Gene map of the Crataegus scabrifolia plastid genome. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise. Genes belonging to different functional groups are color coded. The darker gray in the inner circle corresponds to DNA G + C content, while the lighter gray corresponds to A + T content. LSC: large single-copy; SSC: small single-copy; IR: inverted repeat.
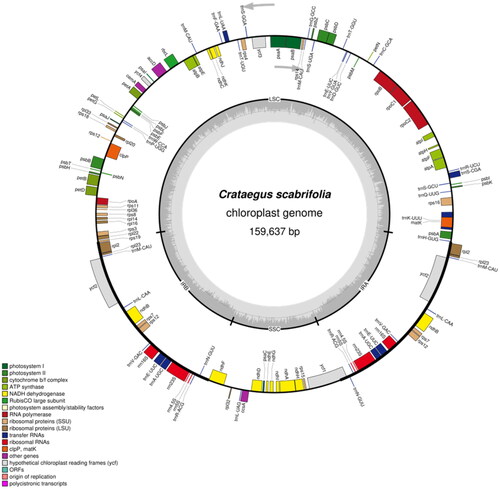
Figure 3. The best maximum-likelihood (ML) phylogenetic tree reconstructed by RAxML ver. 8.0.0 based on complete chloroplast genome sequences from 22 species of Rosaceae, including 9 Crataegus species, Albizia julibrissin, and Cassia fistula as outgroup. The numbers on branches are bootstrap support values from 1,000 replicates. The following sequences were used: Pyrus turcomanica NC 061559.1 (Yang et al. Citation2020); Pyrus georgica NC_061558.1 (Wang et al. Citation2016); Pyrus demetrii NC 061933.1 (Hong et al. Citation2021); Pyrus salicifolia NC 061935.1 (Katayama and Uematsu Citation2005); Pyrus hopeiensis MF521826.1 (Li et al. Citation2021); Pyrus ussuriensis NC 041461.1 (Boby et al. Citation2021); Sorbus insignis NC 051947.1 (Tan et al. Citation2020); Sorbus koehneana NC 063569.1(Li et al. Citation2021); Sorbus rehderiana OK012001.1(Ma et al. Citation2007); Malus ombrophila MW115598.1 (Lu et al. Citation2022); Malus transitoria MK098838.1 (Lu et al. Citation2022); Aronia arbutifolia NC 045391.1 (Taheri et al. Citation2013); Aronia melanocarpa MT527725.1 (Jurikova et al. Citation2017); Crataegus mollis NC 062346.1(Cha et al. Citation2018); Crataegus laevigata NC 062347.1 (Nguyen et al. Citation2021); Crataegus rhipidophylla NC 062345.1 (Żurek et al. Citation2021); Crataegus cuneata NC 058896.1 (Cui et al. Citation2022); Crataegus pinnatifida var. major MW653326.1 (Pang et al. Citation2021); Crataegus pinnatifida MW653325.1 (Guo et al. Citation2021); Crataegus bretschneideri MW963339.1(Zheng et al. Citation2021); Crataegus hupehensis NC 054155.1 (Hu et al. Citation2021); Albizia julibrissin NC 058305.1 (Zhang et al. Citation2021); Cassia fistula ON099431.1 (Sharma et al. Citation2021).

Supplemental Material
Download MS Word (249.7 KB)Data availability statement
The data that newly obtained at this study are available in the NCBI under accession number of OP021659 (https://www.ncbi.nlm.nih.gov/nuccore/OP021659). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA860797, SRR20339770, and SAMN29862852, respectively.
