Figures & data
Figure 1. (a) The leaves of H. pubescens. (b) The follicles of H. pubescens. The leaf blade of this species is ovate or elliptic, with 10–15 pairs of lateral veins, and the follicles of this species are linear, with white, punctate lenticels. The photos of H. pubescens were taken by the authors in Xishuangbanna county, Yunnan Province, China.
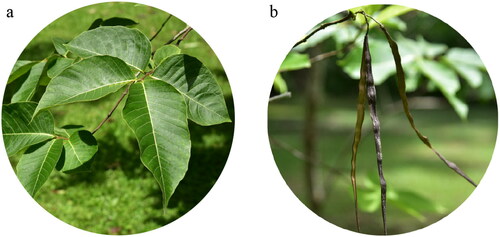
Figure 2. Chloroplast reference genome of Holarrhena pubescens. This figure has six circles from the center to the outside. The first circle shows the forward and reverse repeats connected with the red and green arcs, respectively. The second circle and the third circle show tandem repeats and microsatellite sequences marked with short strips, respectively. The fourth circle indicates the position of LSC, SSC, IRA, and IRB, and the fifth circle indicates the GC content. The genes outside the sixth circle are transcribed counterclockwise, while the genes inside are transcribed clockwise. Genes belonging to different functional groups are color coded, as shown in the left corner.
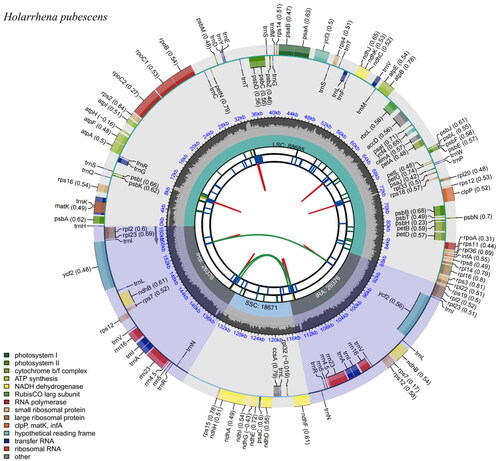
Figure 3. Phylogenetic analysis of 12 species and one taxon as outgroups based on complete protein coding genes sequences by RAxML, bootstrap support value near the branch. The following sequences were used: MK783315 Trachelospermum jasminoides (Wang et al. Citation2019), NC_056322 Chonemorpha megacalyx (Deng et al. Citation2021), MG963251 Beaumontia murtonii (Fishbein et al. Citation2018), GWHBKKE01000000 Holarrhena pubescens, NC_025656 Nerium oleander (Straub et al. Citation2014), NC_054286 Wrightia laevis (Li et al. Citation2020), NC_033354 Carissa macrocarpa (Jo et al. Citation2017), NC_051546 Cerbera manghas (Liao et al. Citation2020), NC_047244 Rauvolfia serpentina (Zhang et al. Citation2021), NC_046841 Rauvolfia verticillata (Chen et al. Citation2019), NC_021423 Catharanthus roseus (Ku et al. Citation2013), NC_065203 Vinca major (Hendrian Citation2007), and NC_027442 Gentiana crassicaulis (Ni et al. Citation2017).

Supplemental Material
Download MS Word (1,012.5 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in the Genome Warehouse of NGDC at https://ngdc.cncb.ac.cn/gwh under the accession number: GWHBKKE01000000. The associated BioProject, GSA, and Bio-Sample numbers are PRJCA011728, CRA008103, and SAMC873599, respectively.
