Figures & data
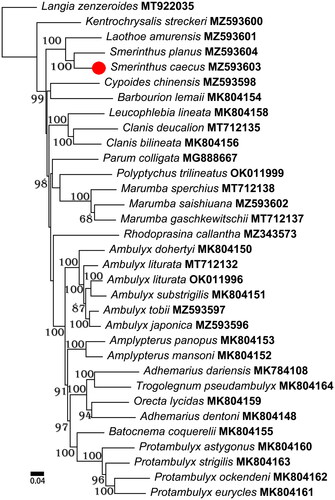
Figure 1. Phylogenetic relationships within Smerinthinae based on the sequences of 13 protein-coding genes analyzed using maximum-likelihood methods. The values of an ultrafast bootstrap (UFB) of 1000 replications are shown on the nodes. The following records were used: ‘Adhemarius dariensis MK784108, Adhemarius dentoni MK804148, Ambulyx dohertyi MK804150, Ambulyx substrigilis MK804151, Amplypterus mansoni MK804152, Amplypterus panopus MK804153, Barbourion lemaii MK804154, Batocnema coquerelii MK804155, Clanis bilineata MK804156, Leucophlebia lineata MK804158, Orecta lycidas MK804159, Protambulyx astygonus MK804160, Protambulyx eurycles MK804161, Protambulyx ockendeni MK804162, Protambulyx strigilis MK804163, and Trogolegnum pseudambulyx MK804164 (Timmermans et al. Citation2019); Ambulyx liturata MT712132, Clanis deucalion MT712135, Langia zenzeroides MT922035, Marumba gaschkewitschii MT712137, and Marumba sperchius MT712138 (Wang et al. Citation2021); Kentrochrysalis streckeri MZ593600 (Huang et al. Citation2022); Smerinthus planus MZ593604 (Meng et al. Citation2022a); Ambulyx tobii MZ593597 (Meng et al. Citation2022b); Laothoe amurensis MZ593601 (Sun et al. Citation2022); Smerinthus caecus MZ593603 (this study); Ambulyx japonica MZ593596, Ambulyx liturata OK011996, Cypoides chinensis MZ593598, Marumba saishiuana MZ593602, Parum colligata MG888667, Polyptychus trilineatus OK011999, and Rhodoprasina callantha MZ343573 (Unpublished)’.
Data availability statement
The data supporting this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MZ593603. The associated BioProject, Bio-Sample numbers, and SRA are PRJNA752517, SAMN20600166, and SRR15358143, respectively.
