Figures & data
Figure 1. A reference image of the sequenced Bassarona dunya collected from Ayer Hitam Forest Reserve, Johor, Malaysia.

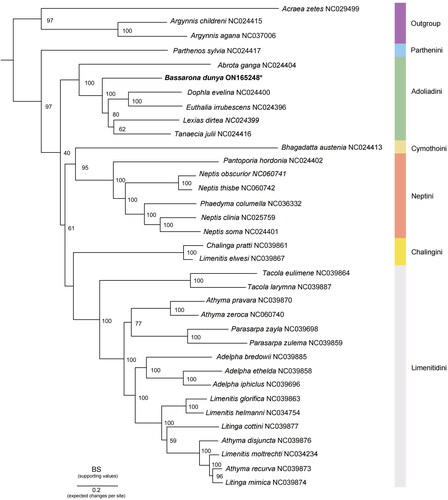
Figure 3. Phylogenetic analysis of B. dunya sequenced in this study, as indicated with an asterisk, and other 34 Nymphalidae mitogenomes. The node values represent the bootstrap of ultrafast 5000 replicates (BS), whereas each color indicates the tribes in Limenitidinae. Outgroup: Acraea zetes (NC029499) (Timmermans et al. Citation2016); Argynnis sagana (Liu et al. Citation2018); Argynnis childreni (NC024415) (Wu et al. 2014). Ingroup: Abrota ganga (NC024404), Euthalia irrubescens (NC024396), Lexias dirtea (NC024399), Dophla evelina (NC024400), Tanaecia julii (NC024416), Bhagadatta austenia (NC024413), Parthenos sylvia (NC024417), Neptis soma (NC024401), and Pantoporia hordonia (NC024402) (Wu et al. 2014); Adelpha bredowii (NC039885), Adelpha ethelda (NC039858), Adelpha iphiclus (NC039696), Athyma pravara (NC039870), Athyma recurva (NC039873), Athyma disjuncta (NC039876), Limenitis glorifica (NC039863), Litinga mimica (NC039874), Litinga cottini (NC039877), Parasarpa zulema (NC039859), Parasarpa zayla (NC039698), Tacola larymna (NC039887), Tacola eulimene (NC039864), Chalinga pratti (NC039861), and Limenitis elwesi (NC039867) (Wu et al. Citation2019); Athyma zeroca (NC060740), Neptis obscurior (NC060741), and Neptis thisbe (NC060742) (Liu et al. Citation2021); Neptis clinia (NC025759) (Tang, et al. Citation2014); Phaedyma columella (NC036332) (Chen et al. Citation2018); unpublished: Limenitis helmanni (NC034754) and Limenitis moltrechti (NC034234); this study: Bassarona dunya (ON165248).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON165248. The associated BioProject, SRA, and BioSample numbers are PRJNA753627, SRR15422669, and SAMN20720549, respectively.